|
|
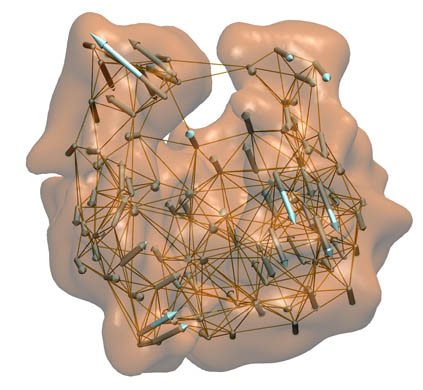
ModeHunter is
a Python-based program package for the normal mode analysis of coarse
grained elastic networks. The package includes the classic
carbon-alpha elastic network model (ENM) , the bend-twist-stretch (BTS) algorithm for
coarse networks from our Situs package, and various useful tools for atomic structure refinement, analysis and
animation.
|
| New in 2023 and 2024: To
prepare for papers on the use of normal modes for refining
AlphaFold2 models, we added in version 1.4 a new tool, elforge.py, for flexible fitting of atomic structures to cryo-EM maps. A related tool, eldecoy.py, was added in version 1.3 for decoy generation
using normal modes. Another tool, traject.py, was also added for
creating animations (trajectories) of mode superpositions. All
ModeHunter tools are now compatible with Python 3. |
|
|

